Whole-genome sequencing (WGS) has revolutionized the field of genomics, enabling scientists to explore the complete genetic makeup of individuals with unprecedented detail. By providing a comprehensive view of the genome, WGS has become a critical tool in research, disease diagnosis, and personalized medicine. However, the accuracy of WGS depends not only on the sequencing technology itself but also on library quality.
Greg Endress Vice President of Innovations Covaris
In this Innovation Spotlight, Greg Endress, vice president of innovations at Covaris, explains how emerging tools and methods help researchers improve library preparation and achieve more accurate sequencing results.
How are scientists currently using WGS in the laboratory and clinic?
WGS is currently utilized as an indispensable methodology in both research and clinical environments. Within the laboratory setting, it is fundamental to large-scale population studies, such as the All of Us Research Program, which seeks to elucidate the genetic underpinnings of various diseases. In clinical practice, WGS is not only vital for the diagnosis of hereditary conditions but is also increasingly integral to precision oncology, where it facilitates the creation of detailed tumor-specific genomic profiles. These profiles provide critical insights that inform tailored therapeutic strategies, thereby advancing personalized medicine by translating complex genomic data into actionable clinical outcomes.
What is the purpose of library preparation in next-generation sequencing (NGS) workflows, and what are the common challenges researchers face during this process?
The primary purpose of NGS library preparation is to convert native DNA molecules into a state suitable for high-throughput sequencing. This intricate, multi-step process entails the systematic fragmentation of DNA to a consistent and optimal size, followed by enzymatic repair of the fragment ends and ligation of unique adapter sequences. These adapters are essential for the sequencing instrumentation to properly identify, cluster, and sequence each molecule. A meticulously executed library preparation protocol is foundational for ensuring the efficient utilization of the sequencer and for generating high-quality, accurate data.
A principal challenge inherent to library preparation is the introduction of methodological variability. Numerous workflows, particularly those reliant upon enzymatic fragmentation, are susceptible to inconsistent DNA fragment sizes, sample-specific biases, and batch-to-batch effects. Such variability can lead to non-uniform genome coverage and diminished data quality. As a result, laboratories are often required to implement labor-intensive and costly quality control measures. These procedural necessities extend turnaround times, escalate operational expenditures, and have the potential to compromise the integrity of the final dataset.
Why is consistent DNA fragmentation important for sequencing accuracy and data quality, and what methods have scientists traditionally used to fragment DNA samples?
Consistent DNA fragmentation is a fundamental prerequisite for generating high-fidelity sequencing data. A uniform distribution of fragment sizes ensures even coverage across the entire genome without sequence bias, thereby preventing data gaps in regions of interest. This uniformity is particularly critical for achieving high-precision variant detection, especially for the identification of small insertions and deletions that are often of significant interest in clinical research. Conversely, inconsistent fragmentation introduces systemic biases that can confound data interpretation and diminish the reliability of the results. Ultimately, consistency in fragmentation enhances the overall efficiency of the sequencing process and improves data integrity.
Traditionally, scientists have employed either mechanical shearing or enzymatic digestion to fragment DNA. While widely used, enzymatic fragmentation methods present several significant problems. These approaches are known to introduce sample-specific biases, particularly in regions with high GC or AT content, and can suffer from batch-to-batch variability between enzyme lots. This inconsistency leads to variable fragment sizes and lower library conversion rates, resulting in extensive and costly quality control procedures to ensure data integrity. Such issues compromise the reproducibility of experiments and can skew the final sequencing results.
What innovative library preparation kits are helping researchers overcome the common WGS challenges?
The Covaris truCOVER® WGS PCR-free Library Prep Kit represents a comprehensive, integrated solution engineered to mitigate the principal challenges associated with library preparation. It uniquely combines the proprietary Adaptive Focused Acoustics® (AFA®) technology, which provides superior mechanical DNA shearing, with a highly efficient, single-vessel workflow. This synergy yields consistently sized DNA fragments and maximizes the conversion efficiency of DNA into sequence-ready libraries. Consequently, the kit facilitates reliable mass-based library pooling, often obviating the need for qPCR, while reducing turnaround time by as much as 30 percent and ensuring the generation of high-quality data from diverse sample types.
The truCOVER Amplification Kit is an ancillary module designed for NGS library preparation involving low-input DNA samples, typically ranging from 0.1 to 50 nanograms. Its primary function is to increase the quantity of library DNA to a level sufficient for sequencing, which is critical when working with precious or limited starting material from sources such as formalin-fixed paraffin-embedded tissues. Covaris engineered the kit to perform this amplification with minimal bias, ensuring that the resulting data accurately reflects the original sample’s genomic profile without skewing due to factors such as GC content. This enables researchers to obtain high-quality sequencing data from challenging samples.
What is AFA, and what are the advantages of using this technology for DNA fragmentation?
AFA is a proprietary Covaris technology that enables highly precise and reproducible mechanical DNA shearing. The technology utilizes controlled bursts of high-frequency acoustic energy focused into a discrete zone within the sample vessel. This non-contact process creates controlled cavitation, resulting in hydrodynamic shear forces that fragment biomolecules.
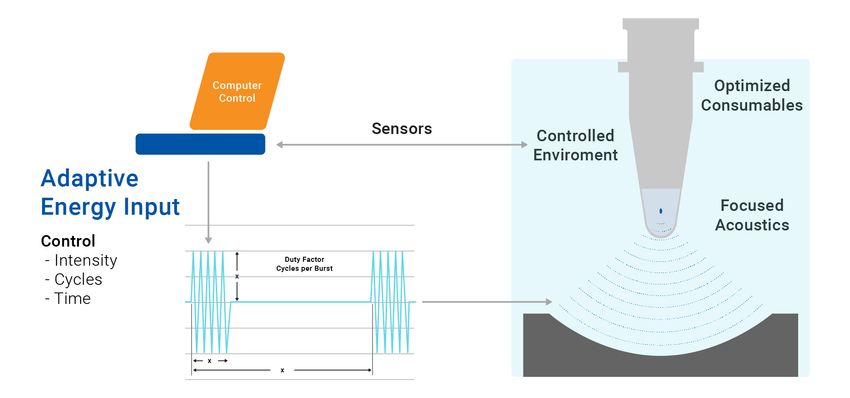
Adaptive Focused Acoustics® (AFA®) technology uses precise high-frequency acoustic energy to efficiently and consistently fragment DNA, ensuring uniform shearing for high-quality results.
Covaris
Because the fragmentation process is highly controlled, AFA delivers exceptional precision and reproducibility, yielding a tight and uniform fragment size distribution. Unlike enzymatic methods, AFA is an unbiased physical process, preventing fragmentation preferences in GC- or AT-rich genomic regions and eliminating batch-to-batch variability. This process enhances data integrity and ensures more uniform genome coverage. The controlled, non-contact nature of AFA also improves workflow robustness across diverse sample types, solidifying its status as the gold standard for high-quality DNA fragmentation.
What are the benefits of using the truCOVER Library Prep Kits over other library preparation methods?
The truCOVER Library Prep Kits provide several distinct advantages over alternative methods. First, by employing AFA technology, they facilitate unbiased and highly reproducible DNA fragmentation, resulting in more uniform genomic coverage and higher confidence in variant calling. Second, the streamlined, single-vessel workflow is designed to minimize sample transfers and manual intervention, which contributes to a reduction in overall turnaround time and avoids sample loss. Finally, the combination of higher library conversion efficiency and exceptional consistency enables accurate mass-based pooling, which frequently eliminates the requirement for expensive qPCR-based quantification and thus lowers the total per-sample cost.
How might innovations like these kits affect the future of genomics research and personalized medicine?
Innovations such as the truCOVER kits serve as catalysts for the advancement of genomics research and personalized medicine. By alleviating critical bottlenecks related to time, cost, and methodological variability, these technologies enhance the accessibility and scalability of high-quality WGS. The resulting standardization is crucial for conducting robust, large-scale population studies and for the reliable detection of clinically significant variants. By providing a more rapid and cost-effective pathway to high-confidence data, this technology empowers researchers and clinicians to more effectively translate genomic information into actionable insights and, ultimately, improve patient outcomes.