Researchers at Baylor College of Medicine have developed a new approach to isolating circulating tumor cells (CTCs) from liquid biopsies of patients with triple-negative breast cancer (TNBC). Through the identification of four new cell surface markers specific to TNBC cells, this work could address a major limitation that has historically prevented the use of CTCs in the diagnosis and monitoring of TNBC.
“The new markers detected cells that standard methods missed,” said Chonghui Cheng, MD, PhD, professor of molecular and human genetics at the Lester and Sue Smith Breast Center at the Baylor College of Medicine and senior author of the study, published today in Cancer Research Communications. “When the four new markers were combined, detection improved substantially. Importantly, the new markers on CTCs showed very little overlap with markers on normal blood cells, reducing the risk of false positives.”
Triple-negative breast cancer is the most aggressive form of breast cancer, characterized by fast progression and early metastasis. Despite major advances in precision oncology, there is still a significant lack of targeted therapies for TNBC, resulting in poor outcomes for patients with this diagnosis.
Capturing CTCs from blood samples offers a minimally invasive option to monitor cancer progression and metastasis, but their scarcity makes them difficult to isolate. Typically, CTCs are captured from a blood draw using antibodies that target cell surface markers unique to tumor cells. However, commonly used markers favor the selection of epithelial CTCs over the mesenchymal cancer cells found in TNBC tumors.
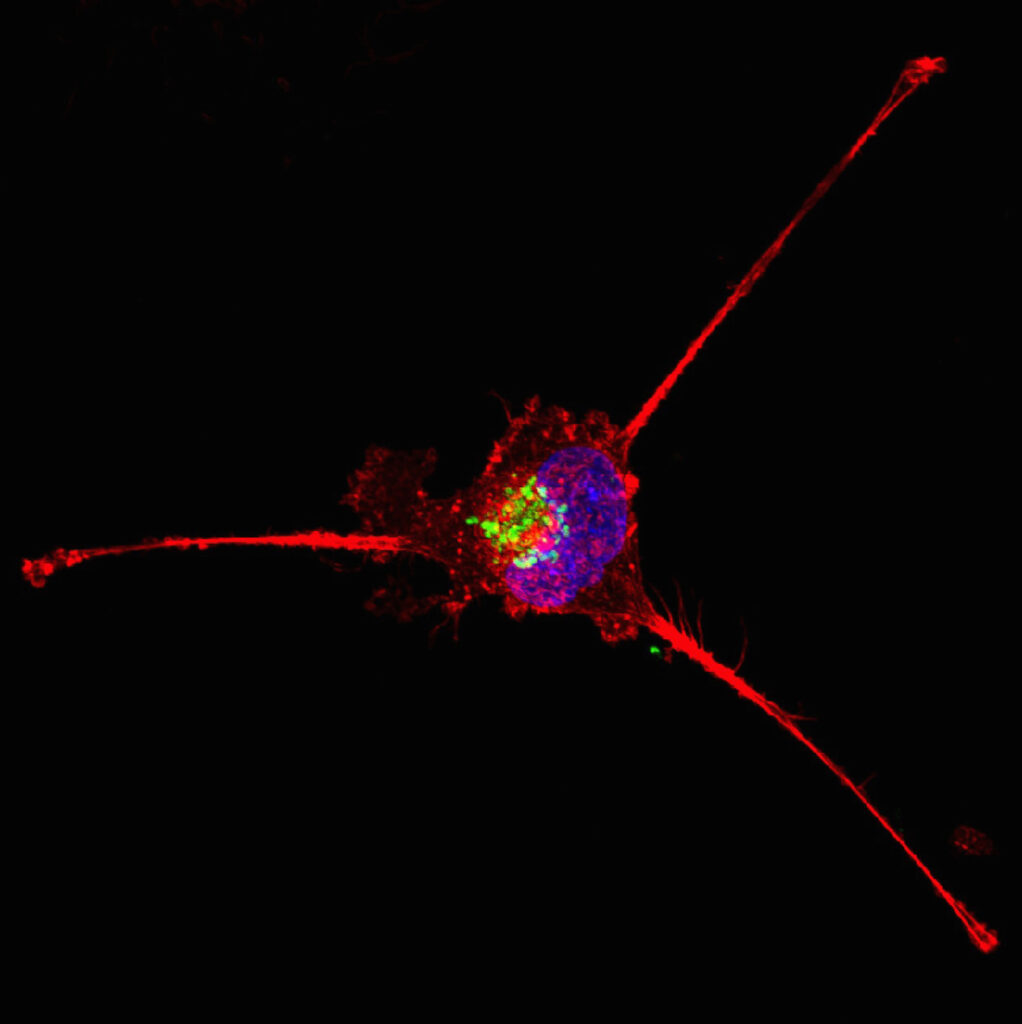
Using a mouse model of TNBC, Cheng and colleagues captured live CTCs and analyzed them using single-cell RNA sequencing. This allowed them to identify four key cell surface proteins found on TNBC CTCs but not on normal blood cells: AHNAK2, CAVIN1, ODR4 and TRIML2.
A combination of antibodies against these four markers significantly improved CTC detection rates in multiple mouse models of TNBC as well as in patient samples. Combining these markers with traditional CTC surface markers further enhanced the sensitivity of the detection method, allowing researchers to identify diverse CTC populations in the bloodstream while preserving RNA quality for single-cell sequencing studies.
“We were excited with the results with blood from patients with metastatic TNBC,” said Cheng. “In these patients, tumor cells were frequently undetectable using standard markers but became clearly visible when we applied the new marker combination.”
This research provides a new path to studying CTCs during disease progression and metastasis, enabling the identification of novel targets for the development of prognostic tools and therapeutic targets for a type of cancer that is notably hard to treat. In a clinical setting, reliable detection of TNBC CTCs could also help physicians monitor patients over time and personalize treatment to their unique needs.
“Another exciting finding is that the newly identified markers are also expressed in other cancer types, suggesting that this strategy could improve CTC detection across multiple cancers,” said Cheng.